Biochemistry
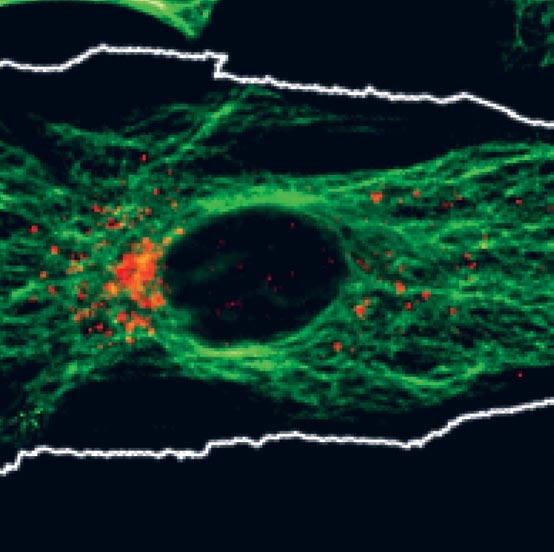
Human cells labeled with antibodies to detect microtubules (green) and mannose 6-phosphate receptors (red).
The future of Biochemistry is to understand how molecules direct all cellular processes, from gene expression to cellular organization to tissue formation and organogenesis. This begins with an understanding of how proteins fold and how enzymes work. But cells are not bags of enzymes; rather, biochemical processes are compartmentalized. For example, signal transduction involves signaling complexes held together by scaffolds, anchors and adaptor proteins. These macromolecular assemblies must be studied in a functional sense to fully understand cell regulation. Biochemical processes are also ordered in relation to the three dimensional organization of a cell. How do cells achieve their polarity, and what molecular constituents drive their ordered division? Now that we can analyze the expression of entire genomes in a single experiment, what kinds of questions can we ask that could never have been asked before?
Students in our department are given opportunities to investigate the biochemical and structural basis of molecular, cellular and developmental processes at a variety of levels of detail and in a variety of organisms ranging from bacteria to mammalian cells. Ongoing research includes studies of protein and RNA folding and catalysis by protein enzymes and ribozymes; DNA replication and recombination; gene regulation; cell motility and molecular motors; protein targeting and transport in the cell; cell adhesion and recognition; intercellular signaling; methods of isolating, and analyzing and altering genes and entire genomes.
The department’s graduate program emphasizes research training and continues to spawn leading researchers in academia and biotechnology. Students design their own programs of study to best suit their educational goals and select the research group for their thesis following research rotations and in consultation with their graduate advisor and other faculty. Concurrent with thesis research, students formulate original research proposals, take classes in biochemistry, structural biology, cell biology and genetics, serve as teaching assistants, attend a journal club with presentations by faculty, students and postdoctoral fellows, and attend seminars by outside speakers. Annual department-wide retreats serve as a forum for presentations by all research groups. Laboratory rooms and equipment are shared by all research groups to promote interactions and exchange of ideas between students, postdoctoral fellows and faculty.
For more information contact:
Joella Mesa
Department of Biochemistry
Beckman Center, B400
Stanford, CA 94305-5307
(650) 725-9058
(650) 723-6783 (fax)
joella.mesa@stanford.edu
http://biochem.stanford.edu/
Faculty and their Research Interests
Steven Artandi is interested in unraveling the molecular and cellular mechanisms according to which telomeres and telomerase modulate stem cell function and carcinogenesis. http://www.stanford.edu/group/artandi/
Phil Beachy My lab studies the function of Hedgehog proteins and other extracellular signals in morphogenesis, and in repair and regeneration of tissue injury. We study how the distribution of such signals in tissues is regulated, how cells perceive and respond to distinct concentrations of signals, and how such signaling pathways arose in evolution. We also study the normal roles of such signals in the physiology of stem and progenitor cells and the abnormal roles of such signaling pathways in the formation and expansion of cancer stem cells.
http://cancer.stanford.edu/features/faculty/beachy.html
Patrick Brown is developing and applying genomic approaches to map global gene expression programs and to understand the architecture and the detailed molecular mechanisms of their regulation. His lab is also exploring new approaches to the classification, detection, diagnosis and treatment of human diseases, developing ex vivo culture systems for reproducing human tissue microenvironments, and systematically investigating the microbial ecology of the human body.
http://cmgm.stanford.edu/pbrown/Pat_Brown_Lab_Home_Page/Home.html
Gilbert Chu studies non-homologous end joining, the pathway that repairs double-strand breaks created by ionizing radiation and V(D)J recombination. The lab is elucidating the mechanism for the end joining reaction, and exploring methods for disrupting the pathway in breast and prostate cancer cells to make them hypersensitive to chemotherapy and radiation therapy.
http://cmgm.stanford.edu/~chu/
Rhiju Das strives to predict how sequence codes for structure in proteins, nucleic acids, and heteropolymers whose folds have yet to be explored. The Das group uses new computational and experimental tools to tackle the ab initio modeling of protein and RNA folds, the high-throughput structure mapping of riboswitches and random RNAs, and the crowd-sourced design of nucleic acid devices.
http://www.stanford.edu/~rhiju/
Ronald Davis is devising new technologies and instrumentation toward rapid whole genome sequencing, expression, proteomics, and functional analysis. These novel approaches are being applied to yeast, humans, plants and various pathogens.
http://med.stanford.edu/sgtc/
James Ferrell, Jr. is studying mitosis and meiosis in Xenopus embryos and human cell lines. His lab is particularly interested in trying to understand how complex biochemical behaviors like switching and oscillations emerge out of small networks of regulatory proteins.
http://www.stanford.edu/group/ferrelllab/
Pehr Harbury. The Harbury lab studies the molecular structure of living systems, and explores strategies to engineer molecules for novel functions. We aim to harness biological paradigms such as evolution and the central dogma as tools for scientific inquiry: examples include (1) recapitulating billions of years of small-molecule evolution in a test tube in order to craft "designer" small molecules; (2) globally rewiring the genetic code to enable visualization of protein structure/dynamics "in action" in complex mixtures; and (3) computationally designing protein active sites.
http://cmgm.stanford.edu/biochem/harbury/
Dan Herschlag is studying the fundamental properties of RNA and protein catalysis and of RNA folding, using a wide array of techniques including single molecule fluorescence, X-ray crystallography, NMR, EPR, vibrational spectroscopy, small angle X-ray scattering, and pre-steady state kinetics. Fundamental principles are also applied on a genomic level to understand RNA structure and dynamics in vivo and the control of RNA processing in gene expression.
http://cmgm.stanford.edu/biochem/herschlag/
KC Huang is trying to understand the relationship between cell shape and behavior in bacteria. His lab uses a combination of computational physics-based models and approaches from experimental evolution and synthetic biology to control morphogenesis and cellular organization.
http://whatislife.stanford.edu
Chaitan Khosla studies the biosynthesis of polyketide antibiotics with the concomitant goal of engineering polyketide synthases to make new bioactive natural products. He also seeks to understand the biochemistry of Celiac Sprue, an autoimmune disease of the small intestine, and to translate these insights into therapeutic solutions for this unmet medical need.
http://www.stanford.edu/group/khosla/Home.html
Mark Krasnow is using genetic, genomic, and biochemical approaches to identify lung stem cells and the molecular program of lung development, and how they go awry in lung cancer and other diseases. He is also mapping the respiratory control circuit, seeking to identify the cellular and molecular basis of breathing, oxygen and carbon dioxide sensing, and breathing arrhythmias such as Sudden Infant Death Syndrome and sleep apnea.
http://cmgm.stanford.edu/krasnow/
Sharon Long studies chemical signals and cellular responses in the development of symbiotic N2-fixing root nodules by Rhizobium bacteria and leguminous plants. She and her lab group study differentiation and morphogenesis with approaches including genetics, natural-product and protein biochemistry, transcriptomics, and advanced imaging techniques.
http://cmgm.stanford.edu/biology/long/
Suzanne Pfeffer is interested in the mechanisms by which receptors traffic between membrane-bound compartments in human cells. Her lab studies how Rab GTPases function with other proteins to help transport vesicles form and identify their targets. She is also studying the NPC1 membrane protein that is needed for cellular utilization of LDL cholesterol, and when mutated, causes Niemann Pick Type C disease in humans.
http://pfeffer.stanford.edu/
Rajat Rohatgi is working to elucidate the biochemical and cell biological principles that govern signaling pathways at the intersection between developmental biology and cancer. A particular focus is signal transduction at primary cilia, an antenna-like projection that is a major coordinating center for cell-cell communication.
http://rohatgilab.stanford.edu/
James A. Spudich focuses on the myosin family of molecular motors, their roles in vivo and the mechanism by which they facilitate cell contraction, cytokinesis, and a variety of other forms of cell movement. The Spudich lab continues to develop modern biophysical tools for single molecule analysis, and is now applying these tools to understand the molecular basis of the debilitating human diseases hypertrophic and dilated cardiomyopathies.
http://spudlab.stanford.edu/
Aaron F. Straight is studying the mechanisms of eukaryotic chromosome segregation and cell division. His laboratory studies the assembly and function of the centromere and kinetochore, the higher order organization of eukaryotic chromosomes, and how non-coding RNAs and other epigenetic mechanisms influence chromosome function. The Straight lab uses biochemistry, genetics, microscopy and cell biological approaches to probe chromatin organization and chromosome dynamics. http://straightlab.stanford.edu/
Julie A. Theriot is studying the mechanics and dynamics of cell shape and movement, using a wide variety of cell types as diverse as human leukocytes, fish skin epidermal cells, soil amoebae, bacterial pathogens, and photosynthetic cyanobacteria. Our group combines biophysical, biochemical, and cell biological approaches, using quantitative videomicroscopy and image analysis as primary tools.
http://cmgm.stanford.edu/theriot/
Emeritus Faculty
Robert L. Baldwin is studying the roles of peptide backbone solvation and peptide H-bonds in the energetics of protein folding.
http://rbaldwin.stanford.edu/
Paul Berg Nobel laureate, studied the molecular mechanisms of genetic recombination with a view to improving the efficiency of genomic modifications in mammalian cells and whole animals. He received the Nobel prize for his fundamental studies of nucleic acids that led to the ability to use recombinant DNA.
http://berg-emeritusprofessor.stanford.edu/
Douglas Brutlag is interested in computational methods for predicting gene function and gene regulation from sequence. He was a pioneer in bioinformatics and is involved in teaching Computational Molecular Biology, Genomics and Proteomics courses.
David Hogness is interested in the molecular basis of Drosophila development, including genetic networks that control the larva-to-fly metamorphosis and their activation by the steroid hormone ecdysone via its receptor. He is the father of modern genome analysis and carried out the first successful cloning of a eukaryotic gene.
http://hogness.stanford.edu/
Dale Kaiser is investigating the coordination of multicellular development in a bacterial system. He is trying to understand how intercellular signaling controls cell movement and gene expression.
Robert Lehman studied the mechanism of DNA replication in eukaryotes, with most recent emphasis on the replication of herpes virus DNA.
