Research Summary
Tom Cech and his research group are studying the structure and mechanism of long noncoding RNAs and RNA-protein complexes, including telomerase and complexes that regulate transcription. Their research also focuses on the telomeric DNA-protein complexes that cap the ends of human chromosomes and help regulate telomerase. The overarching goal is to understand the detailed mechanism of these systems and how their misregulation contributes to diseases, including cancer and amyotrophic lateral sclerosis (ALS).
Our lab's early work on catalytic RNAs (ribozymes) helped to establish that RNA is not restricted to being a passive carrier of genetic information, but can have an active role in cellular metabolism. We have now moved out of this RNA world to investigate other long noncoding RNAs, where catalysis is carried out by RNPs (RNA-protein complexes).
Telomerase, an RNP enzyme critical for chromosome end replication, provides the subject for much of our research. Telomerase is a fascinating biochemical system because a large DNA-protein complex (the chromosome end) must interact productively with an RNA-protein complex (telomerase). In addition, telomerase is biologically and medically important; telomerase mutations lead to diseases involving degeneration of highly proliferative tissues, and activation of expression of telomerase in adult human somatic cells is a step toward cancer.
Telomerase Structure and Function
Telomerase was discovered in 1985 by Elizabeth Blackburn and Carol Greider, who also cloned and sequenced its template-containing RNA subunit. The catalytic protein subunit, however, remained elusive. As a postdoctoral fellow in our lab, Joachim Lingner purified telomerase from the ciliated protozoan Euplotes, which has an astoundingly large number of telomeres, and found that it contained a reverse transcriptase subunit. TERT (telomerase reverse transcriptase) was quickly found in other organisms, including budding and fission yeast, Tetrahymena, and humans. In addition to studying the structure and function of the core telomerase RNP, we are interested in accessory proteins that are often species specific. For example, budding yeast telomerases include the Ku heterodimer, which has the unusual property of binding double-stranded DNA nonspecifically and a particular stem loop of telomerase RNA specifically, either in the same site or a mutually exclusive site within Ku.
Chromosome End-Capping Proteins
Cells must distinguish broken chromosome ends, which elicit a DNA-damage response, from the natural ends called telomeres. Telomeric DNA typically consists of multiple repeats of a short sequence, such as TTAGGG in mammals. These repeats are synthesized by telomerase. The DNA repeats recruit proteins that cap off the chromosome ends, preventing the DNA-damage response, and these proteins also regulate telomerase action. Some telomeric proteins bind double-stranded regions of the telomeric DNA, while others bind the single-stranded DNA "tail" at the very end of the chromosome.
Telomeric single-stranded DNA-binding proteins were first identified in ciliated protozoa, facilitated by the extremely large number of minichromosomes present in these organisms. Our lab cloned and sequenced the genes for two Oxytricha nova telomere end-binding proteins (TEBPs) in 1990 and 1991, but for a decade thereafter it was unclear whether humans even had corresponding proteins. Then in 2001, Peter Baumann in our lab identified candidate genes in Schizosaccharomyces pombe and in humans, which were named POT1 (protection of telomeres 1). The deletion of the gene for Pot1 in S. pombe deprotected the telomeres, leading to rapid loss of all telomeric DNA; rare survivors had circularized all three of their chromosomes, thereby circumventing the need for telomeres. X-ray crystal structures of Pot1 protein-telomeric DNA complexes from S. pombe and human by Ming Lei in our lab revealed the molecular basis of sequence-specific DNA recognition, while work in other labs identified a binding partner of Pot1, now called Tpp1. Thus, the ciliate TEBP α-β dimer has a human homolog, POT1-TPP1.
Our initial expectation was that the binding of POT1-TPP1 to a telomeric DNA primer would inhibit telomerase access if the protein complex were at the DNA 3' end, or have no effect if the protein were binding farther from the DNA end. Instead, human POT1-TPP1 acts as a recruitment and processivity factor, promoting telomerase-telomere binding and stimulating telomerase to synthesize multiple DNA repeats before dissociation. Our current research is aimed at answering the following questions: What is the exact molecular interaction between telomerase and TPP1? Can useful inhibitors of the telomerase-TPP1 interaction be identified, and might they even provide lead compounds to develop anticancer pharmaceuticals?
Long Noncoding RNAs Regulating Transcription
While previous research has emphasized the specific binding of the histone methyltransferase PRC2 to XIST RNA, required for X-chromosome inactivation in females, and to HOTAIR, which regulates expression of the developmentally important HOX genes, our work has revealed widespread promiscuous binding of PRC2 to a large number of RNAs, both in vitro and in vivo. Our genome-wide experiments support a model in which promiscuous binding of PRC2 to RNA allows it to maintain epigenetically silenced chromatin while it ignores genes enriched in marks of active chromatin. Our PRC2 research integrates genome-wide analysis with biochemical and biophysical approaches.
Recently many long noncoding RNPs have been implicated in transcriptional regulation in mammals. We study Polycomb Repressive Complex 2 (PRC2) and Fused in Sarcoma (FUS), both of which have been shown to bind noncoding RNAs that can then help recruit them to chromatin.
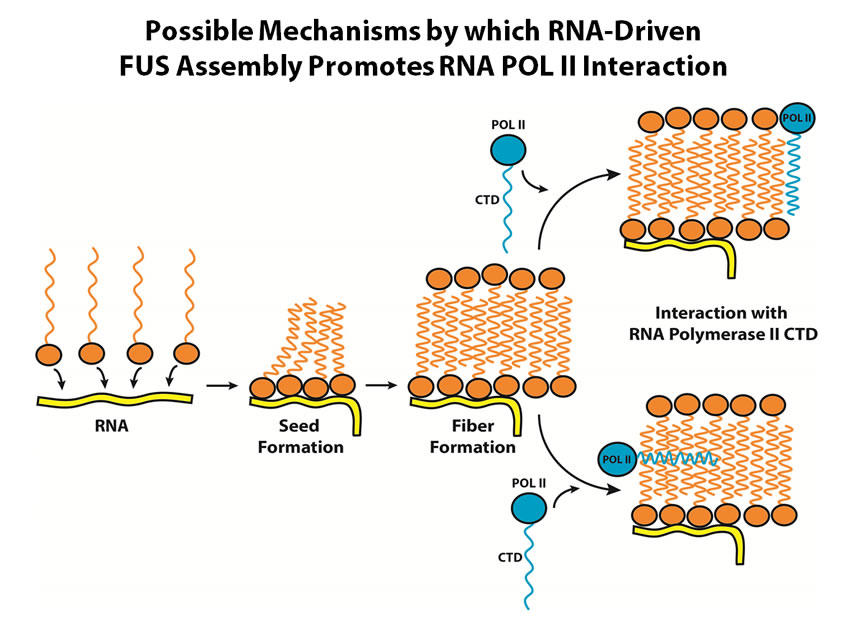
FUS is an abundant nuclear RNA-binding protein associated with two types of disease. Rearrangements of the FUS gene are associated with sarcoma, and localized mutations can cause the neurodegenerative disease ALS. We began working on FUS expecting it to bind specific RNA sequence/structure motifs, but our data have driven us to consider FUS as a more general RNA-binding protein. RNA can provide a platform for cooperative binding of multiple FUS proteins, thereby nucleating the assembly of FUS fibers involving two types of low-sequence-complexity or "prion-like" domains. These assemblies have a positive role in transcriptional regulation in normal cells, but when deregulated they can form the aggregates seen in motor neurons of ALS patients.
Research described above includes projects supported in part by the National Institutes of Health.
As of May 15, 2014