-
 Pma1-mCherry and Vma1-GFP localization in mitotic cells.
Pma1-mCherry and Vma1-GFP localization in mitotic cells.
Image courtesy of M. Eastwood, Fred Hutch and M. Meneghini, University of Toronto. -
 CCCP-induced decrease of mitochondrial membrane potential (below) or control treatment (above) as measured by MitoLoc.
CCCP-induced decrease of mitochondrial membrane potential (below) or control treatment (above) as measured by MitoLoc.
Image courtesy of Dr. Jakob Vowinckel, Ralser Lab, University of Cambridge. -
 Redistribution of Msn5 pools from the nucleus to the cytoplasm upon glucose deprivation.
Redistribution of Msn5 pools from the nucleus to the cytoplasm upon glucose deprivation.
Image courtesy of H. Huang and A. Hopper, Ohio State University. -
 Floccule of yeast rho0 cells expressing PTS1-GFP as a peroxisomal marker, stained with calcofluor white.
Floccule of yeast rho0 cells expressing PTS1-GFP as a peroxisomal marker, stained with calcofluor white.
Image courtesy of Dr. Jakob Vowinckel, University of Cambridge -
 S. cerevisiae membrane proteins visualized by RFP and GFP.
S. cerevisiae membrane proteins visualized by RFP and GFP.
Image courtesy of Masur. Wikimedia Commons. -
 Peroxisome (red) and mitochondrial (green) fission defects in vps1 fis1 double deletion strain transformed with FIS1.
Peroxisome (red) and mitochondrial (green) fission defects in vps1 fis1 double deletion strain transformed with FIS1.
Image courtesy of S. Lefevre, S. Kumar and I. van der Klei, University of Groningen. -
 Yeast cells expressing TRK1/GFP.
Yeast cells expressing TRK1/GFP.
Image courtesy of V. Zayats and J. Ludwig, Center of Nanobiology and Structural Biology, AV CR. -
 The distribution of mtDNA (green) within the mitochondrial network (red).
The distribution of mtDNA (green) within the mitochondrial network (red).
Image courtesy of Christof Osman and Peter Walter, University of California, San Francisco -
 The distribution of ER exit sites (ERES, green) within the ER (red).
The distribution of ER exit sites (ERES, green) within the ER (red).
Image courtesy of A. Nakano and K. Kurokawa, RIKEN. -
 Cell, actin and nuclear morphology of yeast cells treated with DMSO (left) and poacic acid (right).
Cell, actin and nuclear morphology of yeast cells treated with DMSO (left) and poacic acid (right).
Images courtesy of Hiroki Okada and Yoshikazu Ohya, University of Tokyo. -
 Localization of active Ras in a wild type strain
Localization of active Ras in a wild type strain
Image courtesy of S. Colombo and E. Martegani, University Milano Bicocca -
 Sectored colonies showing loss of silencing at the HML locus
Sectored colonies showing loss of silencing at the HML locus
Image courtesy of Anne Dodson, UC Berkeley -
 Pma1p imaged using the RITE tagging system in mother (green) and daughter cells (red)
Pma1p imaged using the RITE tagging system in mother (green) and daughter cells (red)
Image courtesy of Dan Gottschling Ph.D., Fred Hutchinson Cancer Research Center -
 Lipid droplets in fld1 mutant images by CARS
Lipid droplets in fld1 mutant images by CARS
Image courtesy of Heimo Wolinski, Ph.D. and Sepp D. Kohlwein, Ph.D., University of Graz, Austria -
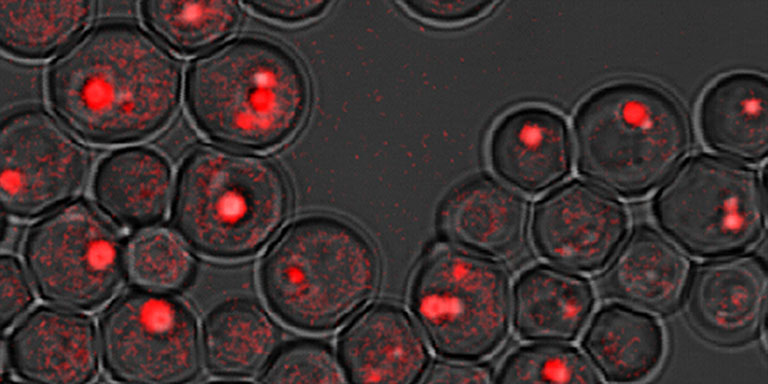
 Fpr3p accumulation in the nucleolus of S. cerevisiae
Fpr3p accumulation in the nucleolus of S. cerevisiae
Image courtesy of Amy MacQueen, Ph.D., Wesleyan University
anti-Fpr3 antibody courtesy of Jeremy Thorner, Ph.D., UC Berkeley -
 San1 strain visualized with FUN and calcofluor white
San1 strain visualized with FUN and calcofluor white
Image courtesy of the Bruschi lab, ICGEB, Trieste, Italy -
 Single MDN1 mRNAs detected by FISH
Single MDN1 mRNAs detected by FISH
Image courtesy of the Zenklusen Lab, Université de Montréal -
 Localization of Ace2-GFP to daughter cell nuclei
Localization of Ace2-GFP to daughter cell nuclei
Image courtesy of Eric Weiss, Ph.D. Northwestern University
About SGD
The Saccharomyces Genome Database (SGD) provides comprehensive integrated biological information for the budding yeast Saccharomyces cerevisiae along with search and analysis tools to explore these data, enabling the discovery of functional relationships between sequence and gene products in fungi and higher organisms.
Upcoming Meetings
Fungal Cell Wall 2015
October 26, 2015 - Paris, France
Abstract deadline: July 2, 2015Genome Informatics
October 28, 2015 - Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
Abstract Deadline: August 14, 2015Cell Biology of Yeasts
November 3, 2015 - Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
Abstract deadline: August 21, 2015Yeast: Products and Discovery 2015 (YPD 15)
December 2, 2015 - Adelaide University, Australia
12th Yeast Lipid Conference
May 20, 2016 - Ghent, Belgium
New & Noteworthy
- Unleashing the Awesome Power of Yeast Transcription
10/07/2015
In the old days, before the internet, planes or even mass publishing, it was hard to spark a quick, worldwide movement. You simply couldn’t reach out to likeminded people who lived far away. Nowadays things are very different. With the advent of social media, it is now trivially easy to spread the word, whether about a revolution or a flash mob. Using Twitter and Facebook, organizers can easily and effectively organize people who live on the... Read... - New SGD Help Video: GO Slim Mapper
10/05/2015
The GO Slim Mapper is a very useful tool that maps specific Gene Ontology (GO) annotations to more general GO terms. This allows you to take a group of genes and bin them into broad categories of function, process, or localization by mapping their GO annotations to broader terms. Watch our new video to get an overview of how the GO Slim Mapper works: ... Read... - The Latest Buzz on Stressed-Out Mitochondria
09/30/2015
Stinging wasps get our attention, and with good reason—getting stung hurts a lot! If you see wasps going into a nest within the walls of your house, you’ll likely try to block their access. But this could backfire: instead of being able to peacefully go into their nest, a swarm of angry wasps could be buzzing around looking for trouble. It might get so bad that you’ll need to call in an exterminator to take care... Read... - A Scientist Sees Transcription
09/23/2015
In the classic Dr. Seuss tale Horton Hears a Who, the elephant Horton thinks he hears voices coming from a speck of dust. He gets into all sorts of trouble over this until all the Whos in Whoville prove they are alive when they all shout at once. Now Horton’s jungle compatriots believe him and Horton can hang out with his new friends. Horton’s companions never get to hear an individual Who. They are not blessed... Read...