Scientists in the early 1900s invented a relatively easy way to measure the force generated by a muscle. They would hang muscle fiber, biopsied from an animal, between two finely calibrated springs and then probe the muscle with an electric shock to make it contract. The stronger the muscle, the more it pulled the springs.

But physiologists soon learned that individual molecules in cells generate the collective force in muscles. And the crude spring experiments couldn’t be applied to these tiny molecules. The challenge of how to measure molecular forces became even more vital as they discovered other molecular motors—molecules in a cell that convert energy to movement. Motors, scientists realized, help sperm swim, pull DNA apart so it can be copied, and carry molecules from one side of the cell to the other. Every directional, nonrandom movement that had been visible under microscopes for centuries was, at some level, due to molecular motors generating force inside cells.
Force is anything that makes an object change its speed, direction, or shape. In the context of cells, forces are required to move molecules. Quantifying these forces gives scientists a way to compare and contrast different molecular motors. Without measurements of force, they were missing a crucial entry in the equations of how cells use energy.
“The bottom line is that the cell is not just a little bag of concentrated reactants. The cell resembles a factory,” says Carlos Bustamante, an HHMI investigator at the University of California, Berkeley. “There are different centers, each specialized in certain functions. Those centers are primarily made up of protein machines. And those machines work as motors that produce torque and movement and force.”
|
WHY MEASURE FORCE? This is research at its purest, done to advance our understanding of the inner workings of the cell. But every molecule that generates or responds to force is one that, when it doesn’t work properly, could lead to cellular malfunction and disease. Understanding the forces at work inside a cell helps scientists discover ways to strengthen or stifle those forces to cause broader changes in a cell. |
Today, thanks to techniques that Bustamante helped bring from physics to biology, scientists can quantify the forces and movements generated by molecular motors. They can precisely measure the force it takes to unfold a protein or unwind a strand of DNA. The innovative methods not only provide fascinating insight into the magnitude of force being generated inside living cells, they also offer ways to change this force and study the consequences. Scientists can play tug of war with a strand of DNA or pull a protein backward along a track to see how the molecules behave under stress.
Shining a Light on Force
In the 1980s, Bustamante worked at the University of New Mexico studying how pieces of DNA moved through gels. When coaxed through the gel by an electric current, fluorescently stained DNA strands moved at different speeds depending on their sizes.
“As I was watching this separation, what became evident to me was how elastic these molecules appeared,” he says. “Sometimes a little piece would get caught and the DNA would stretch out, and then it would snap back like a spring.”
He became curious about the elasticity of DNA and how to measure it. How much force would he need to stretch out a strand like the gel was doing?
In his earliest calculations, Bustamante estimated that he needed a tenth of a piconewton to begin to stretch DNA. “A newton is about the weight of an apple on the surface of the earth,” he says. A piconewton is a millionth of a millionth of that, around the weight of a red blood cell.”
To generate this small amount of force, Bustamante attached one end of a DNA strand to a glass coverslip and the other end to a tiny magnetic bead. Then, he used a second magnet, with a known magnetic strength, to tug on the magnetized end of the DNA strand. He could measure how strong the magnet had to be to stretch the DNA by different amounts. It was the first direct measurement of the elasticity of a strand of DNA and was reported in Science in 1992.
Over the next decade, Bustamante and his colleagues refined the method and brought cutting-edge physics to bear. Instead of using magnetism, their techniques relied on optics, or light. These methods allowed them to make more precise measurements and apply even smaller forces.
If a powerful laser shines through a plastic bead, the light beam is slightly deflected at the bead’s surface. This change in direction of the light beam requires a tiny amount of force. And according to Newton’s third law—for every action there is an equal and opposite reaction—this miniscule amount of force pulls the tiny bead toward the center of the beam. Change the intensity of light, and the amount of force exerted on the bead changes.
Physicist Steven Chu, now the U.S. Secretary of Energy, won the 1997 Nobel Prize in Physics for his quantum physics application of this technique, called optical trapping because it traps a particle in the beam of light. Bustamante was among a handful of scientists who pioneered its use in biology for single-molecule studies.
Page 2 of 4

The small plastic bead used in optical trapping can be attached to a strand of DNA or a protein and pulled using the force generated by the laser beam. To stretch a piece of DNA, Bustamante could attach a plastic bead in place of the magnet, put it under the laser, and slowly move the laser in one direction.
“We knew we were doing experiments that hadn’t been done before,” says Bustamante. “But we came to realize that besides just learning what the elasticity of DNA was, these techniques offered the chance to learn about other interesting things in the cell.”
Suddenly, Bustamante had a way to physically manipulate any molecule that he wanted. He imagined using optical traps to pull proteins apart or drag motors along pieces of DNA. Today, these experiments are reality, and optical trapping is the go-to way for biologists to push and pull on individual molecules to study their behavior.
Measuring Moving Parts
At Yale University, HHMI investigator Anna Pyle studies the shuffling movements of RNA helicases along strands of RNA, the genetic material that translates DNA codes into proteins. As they move, some RNA helicases push other molecules off the RNA. Other helicases are required to unwind double strands of RNA or to recognize foreign RNA brought into a cell by a virus.
“At their core, all these proteins work by opening and closing, shuffling along an RNA strand,” says Pyle. “But that behavior is coupled to all sorts of different functions in the cell.”
Having studied the biochemistry and structure of helicases, Pyle wanted to quantify the force it took for the proteins to move along RNA. In collaboration with Bustamante, she used an optical trap to tug on a helicase as it moved along an RNA strand. As the helicase moved, the optical trap exerted an increasing amount of force on the bead attached to the helicase. The scientists could measure these forces through the laser beam holding the bead in place.
“Getting these numbers on force serves as a real window into basic thermodynamics of these motors,” says Pyle. Biochemists like to think of chemical reactions in terms of equations, she says, and force has been a missing number in those equations. She can now use her initial results to compare the force used by different helicases or to see how a mutation changes the force a helicase can generate, and thus, its function.
Optical trapping experiments by Michelle Wang, an HHMI investigator at Cornell University, upended ideas on how one protein works. In the 1990s, Wang was part of a Princeton University team that used the technique to measure forces of a DNA-based motor protein for the first time. Today, Wang has turned her attention to T7 helicase, a molecular motor that separates double-stranded DNA into two single strands by pulling the DNA through the center of its donut-shaped structure. Other proteins then add nucleotides—the building blocks of DNA—to turn the single strands into two double strands.
T7 helicase can bind two forms of cellular energy. One, called ATP, is the most common currency of energy in cells. Breaking ATP’s chemical bonds releases energy used in many molecular motors. The other form, called dTTP, is both an energy-storing molecule and a DNA nucleotide. Previous experiments with T7 helicase showed that when only ATP was present, the helicase didn’t unwind DNA. But the studies looked at many strands of DNA and helicases at once, averaging how fast the motors moved. Wang wasn’t convinced the collective results told the whole story, since she knew ATP could bind to the helicase.
“When you do an experiment like that, you don’t know the behavior of each molecule and it’s hard to interpret,” says Wang. “It’s like trying to analyze a whole bunch of runners going at different speeds. But instead of measuring the speed of each runner, you measure how long it takes until the last runner crosses the finish line.”
Wang’s team devised optical traps to hold two DNA strands in place, so they could track the unwinding progress of the T7 helicase one molecule at a time. In the presence of dTTP, their associated helicase unwound the DNA at a consistent rate. With only ATP present, the helicase unwound the strands at a faster rate but constantly slipped backward on the DNA, never getting to the end. The group published their work October 6, 2011, in Nature.
Page 3 of 4
“In bulk studies, researchers didn’t get an unwinding signal at all in the presence of ATP,” says Wang. “So we didn’t know this was happening.” She thinks the slippage is an adaptation by the cell to slow unwinding of DNA when there aren’t nucleotides around to quickly bind and pair with each single strand. Since dTTP is a nucleotide that can be incorporated into the strands, its binding to the helicase signals that there are plenty of nucleotides around and the single strands won’t be left hanging.
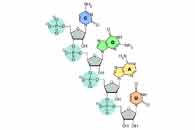
| Helicase T7 encircles a single strand of DNA as it unwinds double-stranded DNA. Each of the six helicase subunits is shown in a different color. The affinity of a subunit to DNA is represented by a small hook: a closed hook for higher affinity and an open hook for lower affinity. When both ATP and dTTP are present, a subunit may bind to either an ATP (lower affinity to DNA, open hook) or a dTTP (higher affinity to DNA, closed hook). The helicase will not slip as long as at least one subunit does not release the DNA. |
Molecular Tug of War
The first applications of optical trapping in biology revolved around DNA and RNA. Since Bustamante had already calculated the elasticity of the molecules, these measurements provided a starting point for other experiments on the forces exerted on or by the nucleic acids. But scientists soon wanted to manipulate proteins on their own—to study how they can alter their complex conformations, unfold, and interact with each other. Optical trapping offered a way to do this physical wrangling.
At the Massachusetts Institute of Technology (MIT), HHMI investigator Tania Baker, as part of a long-term collaboration with HHMI scientific review board member Bob Sauer, runs experiments that are similar to those Michelle Wang set up to study T7 helicase. But Baker studies how one molecular machine, ClpXP, unfolds entire proteins rather than DNA strands.
“Proteins are designed to be really stable in cells, but there are critical times when the cell needs to unfold them,” says Baker. Unfolding a protein inactivates it if the protein is no longer needed, has to cross a membrane, or needs to be remodeled, she explains.
ClpXP, like helicase T7, is ring shaped, and pulls proteins through its center. But it doesn’t always move at a steady rate—some proteins, or parts of proteins, are harder to unfold and cause ClpXP to stall, while other proteins or sections unravel easily, especially once a neighboring bit is unfolded. Baker wanted a way to study the range of speeds at which ClpXP moves as it unwinds different parts of a protein.
So she collaborated with biophysicist Matt Lang, then at MIT and now at Vanderbilt University, to devise an optical trap setup. Two traps held either end of a protein strand in place. As ClpXP unfolded the protein, the strand lengthened—measurable by determining the distance between the traps. So far, she’s shown that the technique works, quantifying the stop–start motion of the unfolding process. Next, she’ll use it to tackle the tougher question of what determines how hard it is for ClpXP to yank protein sections apart.
“We do a lot of biochemistry and a lot of structural studies, and now this is another tool to study this family of enzymes,” says Baker. “One of the things this protein does is create force, so it’s important to study that aspect of it.”
Not all motors in the cell are pulling molecules apart. Some are vehicles, carrying cellular supplies from one location to another. A neuron, for example, has a long process—the axon—that can extend up to one meter. Proteins, membranes, and chemicals must move rapidly from one end of the axon to the other, requiring a molecular motor.
|
|
| Arne Gennerich, a postdoc in the Vale Lab, explains how he uses an optical trapping microscope to analyze dynein. |
In 1985, HHMI investigator Ron Vale of the University of California, San Francisco, discovered kinesin, the molecular motor that transports materials through neurons on filaments called microtubules. In his early experiments, Vale could watch kinesin moving a plastic bead along microtubules under a microscope and later could follow the movement of the motor by single-molecule fluorescence microscopy. But in their natural state, molecular motors of the neuron need to produce a reasonable amount of force to drag their cargos through the dense environment of the cytoplasm. Vale found optical traps to be a useful tool for studying this force.
“It’s like learning how an engine works by studying how it performs under different loads,” says Vale.
|
|
| Watch kinesin as it uses energy from ATP to move stepwise along a microtubule. |
In his latest experiments, optical traps have allowed him to push and pull a single kinesin molecule along microtubules and observe how it responds. Unexpectedly, he found that simply pulling on the kinesin causes it to take regular steps along the microtubule, even in the absence of the chemical energy that it usually needs to produce movement. He also found that he could pull the molecule backward along microtubules, but it takes more force. The difference in the required force provides clues about how kinesin works and how it moves in the correct direction.
The Force of Innovation
While optical traps have answered some questions posed by biologists and given them a way to quantify force in their systems, the method has also led to more questions.
Page 4 of 4
Vale, for instance, now wants to know how kinesin’s structure changes while it’s stepping along microtubules. The atomic details of protein structure can be obtained by x-ray crystallography but are not visible under a light microscope; thus optical traps alone do not provide data on structural changes.
“I’m fascinated by the idea of putting these two worlds of x-ray crystallography and light microscopy together,” says Vale. “What are the real structural changes that are occurring during force generation?”
HHMI investigator Taekjip Ha, at the University of Illinois at Urbana–Champaign, has developed a technique that offers a way to pair structural data with the force control of optical trapping.
In 1996, Ha developed a method to determine the proximity of two fluorescent molecules based on the light they give off. The technique, called fluorescence resonance energy transfer (FRET), had been around for decades, but he showed that it could be used on two single molecules, rather than as an average. The fluorescent tags can be attached to two molecules or two parts of a molecule. As the two tags come closer together or move apart, the fluorescence changes. He uses FRET as a measure of distance, and therefore movement, between any molecules or parts of molecules.
In a test of the method, Ha collaborated with Pyle to uncover details of how one particular helicase—from the hepatitis C virus—unwinds DNA. Its DNA-unwinding function is vital for the virus to make new DNA and infect cells. The scientists attached fluorescent tags to two strands of DNA and attached the strands to optical traps. As the helicase moved along the double strand, separating it, the researchers could observe the unwinding of the DNA, base pair by base pair, as the fluorescent tags got farther apart.
The pair discovered that the helicase unwinds three base pairs at a time, then releases tension in the strand, letting it relax, before unwinding three more. The discovery could help them understand how to block the helicase from helping the virus replicate.
Next, they want to know how much force this unwinding takes. So Ha is combining FRET with experiments measuring force. By measuring how the distance between two parts of a protein changes as a result of force, scientists can get a fuller picture of how unfolding or conformational changes happen.
|
|
| This video shows how the Ha lab is using a combination of high resolution optical trapping with single molecule fluorescence detection. DNA (blue) is tethered between two trapped beads (beads are gray, traps are red). A helicase motor molecule (center) on the DNA is labeled with a single fluorophore (glowing orange) that is excited and detected by a confocal microscope (green). In order for this new instrument to work, the trap and fluorescence excitation lasers have to be rapidly cycled on and off in sequence (called interlacing). This movie shows one complete cycle. |
And Bustamante is now using similar techniques to probe the basics of numerous biological processes, including protein folding. He’s using optical traps and fluorescent tags to see what happens when a protein strand is stretched and then released, allowing it to fold into its preferred conformation.
He’s also applying the method to nucleosomes—clumps of proteins that control the structure of DNA within a chromosome and influence when genes are expressed. He’s already looked at the interaction between polymerases—enzymes that move along DNA strands—and nucleotides. His team discovered that when polymerases encounter a nucleosome, they pause, not having enough force to unravel the DNA from the nucleosome. Instead, the protein waits for the clump to spontaneously unravel. If he can use optical traps to pull apart a fluorescent chromosome, Bustamante says, he can observe its higher-order structure and the forces that proteins within the nucleosomes exert on the nucleotides.
“It’s natural that as optical trapping starts to mature, we now want to combine these techniques with others,” says Bustamante. “I think in the future we will see even more hybrid experiments that combine optical trapping with other methods.”
He’s happy to see his technique mature and change, he says, if it means applying it to more biological questions.
“There are so many unknowns inside the cell,” he says. Optical trapping lets researchers get a physical handle on those unknowns. While scientists can’t reach inside cells and feel for themselves the forces at work, optical trapping has become their hands that work to sense and manipulate these forces.
- 1
- 2
- 3
- 4